Research
Welcome to the world of miniaturized biological screenings. You can explore exciting research projects and the publications of our research group in the Institute of Biological and Chemical Systems at the Karlsruhe Institute of Technology (KIT). Share our fascination about small-scale biology and discover the potential for personalized applications. Join us on this journey of discovery and innovation.
Research Topics
-
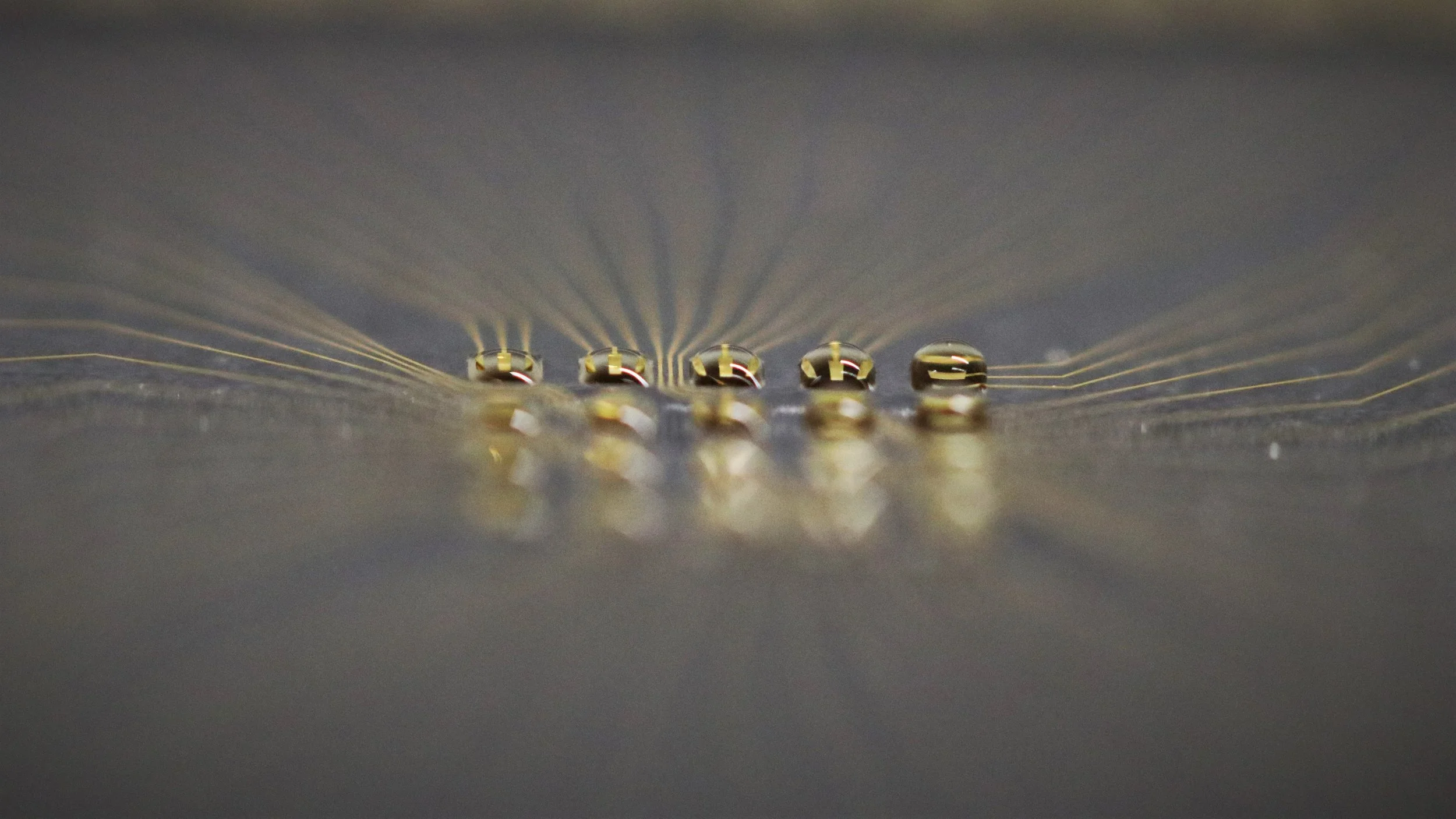
Our research is focused on Droplet Microarray (DMA) Technology, which consists of an array of hydrophilic spots on a superhydrophobic background. This is achieved through chemical modifications of various surfaces and coatings. Due to the extreme difference in wettability between hydrophilic and superhydrophobic areas, it is possible to form arrays of stable nanoliter droplets on a completely planar surface without any physical barriers. These nanoliter droplet arrays offer a novel and miniaturized approach to conducting high-throughput biological screenings on a nanoliter scale, thereby opening up new possibilities in the realm of advanced scientific research and discovery.
-
We develop various workflows and read-out techniques for performing high-class, advanced biological screenings on the DMA platform. Our developed methods include low-input proteomics from the chip, differential gene expression analysis from the chip, label-free real-time monitoring of cell behavior on the chip using electrode arrays, and others. Additionally, we focus on developing workflows and robotics for handling nanoliter volumes, enabling various multiplex and complex workflows necessary for facilitating novel discoveries using DMA technology.
The project in this topic include:
(1) We are involved in the development and investigation of cell behavior in various 3D cell culture models, such as self-assembled spheroids and 3D scaffold-based cell cultures. We explore different types of hydrogel scaffolds for 3D cell culture, accommodating various cell types, including patient-derived cancer cells. To create 3D scaffold structures containing cells on the DMA chip, we employ low-volume dispensing and 3D printing techniques.
(2) Developing protocols and workflows to enable the analysis of differential gene expression in cells treated with drugs on the DMA chip. In this protocol, we lyse the cells, isolate mRNA, and convert it to cDNA in situ. Subsequently, we can utilize the obtained cDNA for either qPCR or mRNAseq analysis. This methodology allows for an in-depth examination of changes in the transcriptome of the cells following drug treatment, in highly miniaturized formats.
(3) Developing and validating novel eDMA (electrode DMA) platform, which is fusion of MEA (micro electrode arrays) and DMA technologies on a single platform. This novel approach will enable lable-free real-time monitoring of cell activity.
(4) Developing workflows enabling analysis of proteome of cells treated with different drugs on DMA chips.
(5) Developing various methods and machines to handle nanoliter volumes on DMA chips including sandwiching and other methods for parallel droplet transfer and sampling, and building fully automated droplet sampler (ANDeS) for sampling the droplets from DMA chip and others.
-
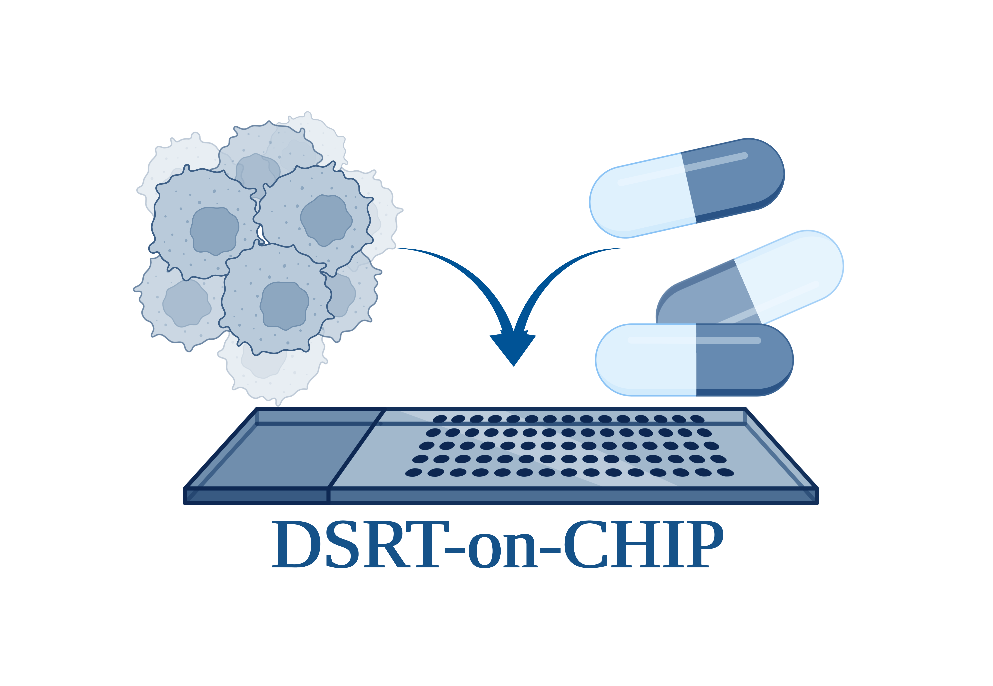
Functional precision medicine through experimentally testing the response of patient-derived cancer cells to anti-cancer therapy in vitro, so-called Drug Sensitivity and Resistance Test (DSRT), has the potential to help identify a suitable therapy for each individual cancer. The main obstacle to establishing this test in clinics is the limited number of patient material, especially for solid types of tumours. This challenge could be addressed by drastic miniaturisation of DSRT on the DMA chips, with possibility for different cell culture models (2D, 3D via spheroids & hydrogels, co-culture), paving the way for the development of DSRT products that eventually can be routinely applied as functional prognostic tests for personalised oncology clinical care.
In our group, we develop protocols for obtaining cells from solid tumor specimens, as well as culturing and treating patient-derived cells on a chip. We work with various types of cancer, currently including blood cancer (CLL), lung cancer (NSCLC), head and neck cancer, and glioma
Publications
Razan El Khaled El Faraj, Shraddha Chakraborty, Meijun Zhou, Morgan Sobol, David Thiele, Lilly M Shatford-Adams, Maximiano Correa Cassal, Anne-Kristin Kaster, Sascha Dietrich, Pavel A Levkin, Anna A Popova (2025) Drug-Induced Differential Gene Expression Analysis on Nanoliter Droplet Microarrays: Enabling Tool for Functional Precision Oncology. Adv. Mat. Technologies DOI: 10.1002/adhm.202401820.
D.D. Kartsev, Urrutia Gómez Joaquin E, Popova A. Anna, Pavel A. Levkin (2025) Droplet Microarrays for Miniaturized and High-Throughput Experiments: Progress and Prospectives. Adv. Mat. Interfaces https://doi.org/10.1002/admi.202400905.
Maryam Salarian, Pavel A. Levkin, Anna A. Popova (2024) Nanoliter Hydrogel Array for Cell Screening and Cell Spheroid Sorting. Adv. Mat. Technologies https://doi.org/10.1002/admt.202401159.
Joaquin E. Urrutia Gómez, Meijun Zhou, Nikolaj K. Mandsberg, Julian A. Serna, Julius von Padberg, Sida Liu, Markus Reischl, Pavel A. Levkin, Anna Popova (2024) Highly Parallel and High-Throughput Nanoliter-Scale Liquid, Cell, and Spheroid Manipulation on Droplet Microarray. Adv. Funct. Mat. https://doi.org/10.1002/adfm.202410355.
Meijun Zhou, Joaquin E. Urrutia Gomez, Nikolaj K. Mandsberg, Sida Liu, Sabine Schmidt, Matthias Meier, Pavel A. Levkin, Heinz-Georg Jahnke, Anna Popova (2024) Electrode Droplet Microarray (eDMA): An Impedance Platform for Label-Free Parallel Monitoring of Cellular Drug Response in Nanoliter Droplets. Adv. Healthc. Mat. https://doi.org/10.1002/adhm.202402046.
Yanchen Wu, Joaquin E. Urrutia Gomez, Hongmin Zhang, Fei Wang, Pavel A. Levkin, Anna A. Popova*, Britta Nestler* (2024) Digital twin of a droplet microarray platform: Evaporation behavior for multiple droplets on patterned chips for cell culture. Droplet https://doi.org/10.1002/dro2.94
Joaquín E. Urrutia Gómez, Razan El Khaled El Faraj, Moritz Braun, Pavel A. Levkin, Anna A. Popova (2023) ANDeS: An automated nanoliter droplet selection and collection device. https://doi.org/10.1016/j.slast.2023.11.002
Marcel P. Schilling, Razan El Khaled El Faraj, Joaquín Eduardo Urrutia Gómez, Steffen J. Sonnentag, Fei Wang, Britta Nestler, Véronique Orian-Rousseau, Anna A. Popova, Pavel A. Levkin, and Markus Reischl (2023) Automated high-throughput image processing as part of the screening platform for personalized oncology. Sci Rep doi: 10.1038/s41598-023-32144-z
Iwohn, Michelle J.; Seifermann, Maximilian; Reiser, Patrick; Höpfner, Julius; El Khaled El Faraj, Razan; Heißler, Stefan; Popova, Anna A.; Levkin, Pavel A (2023). OligoHydrogelArray (OHA) for Parallelized Solid‐Phase Extraction of Oligonucleotides. Adv. Mater. Interfaces https://doi.org/10.1002/admi.202300227
Haijun Cui, Xueyuan Sun, Marcel Schilling, Christel Herold-Mende, Markus Reischl, Pavel A Levkin*, Anna A Popova*, Şevin Turcan* (2023). Repurposing FDA-Approved Drugs for Temozolomideresistant IDH1 Mutant Glioma using High-Throughput Miniaturized Screening on Droplet Microarray Chip. Adv Healthc Mater. PMID: 37162029 DOI: 10.1002/adhm.202300591
Haijun Cui, Tina Tronser, Xianxian Wang, Janine Wesslowski, Gary Davidson, Anna A. Popova,* Pavel A. Levkin (2022). High‐throughput formation of miniaturized cocultures of 2D cell monolayers and 3D cell spheroids using droplet microarray. Droplet e39, https://doi.org/10.1002/dro2.39
Wenxi Lei, Anke Deckers, Charlotte Luchena, Anna Popova, Markus Reischl, Nicole Jung, Stefan Bräse, Thomas Schwartz, Ilga K Krimmelbein, Lutz F Tietze, Pavel A Levkin (2022). Droplet Microarray as a Powerful Platform for Seeking New Antibiotics Against Multidrug‐Resistant Bacteria. Adv Biol. e2200166, doi: 10.1002/adbi.202200166
Y Liu, S Bertels, M Reischl, R Peravali, M Bastmeyer, AA Popova*, Pavel Levkin (2022). Droplet microarray based screening identifies proteins for maintaining pluripotency of hiPSCs. Adv. Healthc. Mater. e2200718, doi: 10.1002/adhm.202200718
Shraddha Chakraborty, Charlotte Luchena, Jonathan J. Elton, Marcel P. Schilling, Markus Reischl, Margaux Roux, Pavel A. Levkin* and Anna A. Popova* (2022). ‘Cells-to-cDNA on Chip’: Phenotypic assessment and gene expression analysis from live cells in nanoliter volumes using droplet microarrays. Adv. Healthcare Mater doi: 10.1002/adhm.202102493
Marcel P. Schilling, Svenja Schmelzer, Joaquin Eduardo Urrutia Gomez, Anna A. Popova, Pavel A. Levkin and Markus Reischl∗ (2021). Grid Screener: A Tool for Automated High-Throughput Screening on Biochemical and Biological Analysis Platforms. IEEE Access DOI: 10.1109/ACCESS.2021.3135709
MP Schilling, L Rettenberger, F Münke, H Cui, AA Popova, PA Levkin, Ralf Mikut, Markus Reischl (2021). Label assistant: A workflow for assisted data annotation in image segmentation tasks. Proc. Workshop Comput. Intell., 211-234, https://doi.org/10.5445/KSP/1000138532
Anna A. Popova,∗ , Markus Reischl, Daniel Kazenmaier, Haijun Cui, Timo Amberger, Pavel A. Levkin∗ (2021). Simple assessment of viability in 2D and 3D cell microarrays using singlestep digital imaging. SLAS Technology doi: https://doi.org/10.1016/j.slast.2021.10.017
Yanxi Liu, Shraddha Chakraborty, Chatrawee Direksilp, Johannes M. Scheiger, Anna A. Popova*, Pavel A. Levkin* (2021). Miniaturized droplet microarray platform enables maintenance of human induced pluripotent stem cell pluripotency. Materials Today Bio doi: https://doi.org/10.1016/j.mtbio.2021.100153
Shraddha Chakraborty, Victor Gourain, Maximilian Benz, Johannes M. Scheiger,c, Pavel A. Levkin* and Anna A. Popova,* (2021). Droplet Microarrays for cell culture: effect of surface properties and nanoliter culture volume on global transcriptomic landscape. Materials Today Bio doi: https://doi.org/10.1016/j.mtbio.2021.10011
Haijun Cui, Xianxian Wang, Janine Wesslowski, Tina Tronser, Jakob Rosenbauer, Alexander Schug, Gary Davidson, Anna A. Popova,* and Pavel A. Levkin* (2021). Assembly of Multi-Spheroid Cellular Architectures by Programmable Droplet Merging. Advanced Materials doi:10.1002/adma.202006434
C RamalloGuevara, D Paulssen, AA Popova, C Hopf, PA Levkin (2021). Fast Nanoliter‐Scale Cell Assays Using Droplet Microarray–Mass Spectrometry Imaging. Adv. Biol. 5 (3), 2000279
Anna A. Popova, Sascha Dietrich, Wolfgang Huber, Markus Reischl, Ravindra Peravali1, and Pavel A. Levkin (2020). Miniaturized Drug Sensitivity and Resistance Test on Patient-Derived Cells Using Droplet-Microarray. SLAS Technology doi: 10.1177/2472630320934432
Popova AA, Levkin PA (2020) Precision Medicine in Oncology: In Vitro Drug Sensitivity and Resistance Test (DSRT) for Selection of Personalized Anticancer Therapy Adv. Therap. DOI:10.1002/adtp.201900100
G Oudeng, M Benz, AA Popova, Y Zhang, C Yi, PA Levkin, M Yang (2020). Droplet microarray based on nanosensing probe patterns for simultaneous detection of multiple HIV retroviral nucleic acids. ACS applied materials & interfaces 12 (50), 55614-55623, https://doi.org/10.1021/acsami.0c16146
Popova AA, Tronser T, Demir K, Kuodyte K, Starkuviene V, Wajda P, Levkin PA (2019) Facile One Step Formation and Screening of Tumor Spheroids Using Droplet‐Microarray Platform Small doi.org/10.1002/smll.201901299
Popova AA, Marcato D, Peravali R, Wehl I, Schepers U, Levkin PA (2018) Fish-Microarray: Miniaturized Platform for Single-Embryo High-Throughput Screenings Adv Funct. Mater. doi.org/10.1002/adfm.201703486
Tronser T, Popova AA, Jaggy M, Bastmeyer M, Levkin PA (2017) Droplet Microarray Based on Patterned Superhydrophobic Surfaces Prevents Stem Cell Differentiation and Enables High-Throughput Stem Cell Screening. Adv Healthc Mater. doi: 10.1002/adhm.201700622.
Tronser T, Popova AA, Levkin PA (2017) Miniaturized platform for high-throughput screening of stem cells. Curr. Opin. Biotechnol. 4: 46: 141-149 doi: 10.1016/j.copbio.2017.03.005.
Gabriella E. Jogia, Tina Tronser, Anna A. Popova and Pavel A. Levkin (2016) Single Cell Analysis using Droplet Microarray Microarrays, 2016, 5(4):28. doi:10.3390/microarrays5040028.
Anna A. Popova, Claire Depew, Katya Manuella, Alexander Trubitsyn, Ravindra Peravali, Jorge Ángel González Ordiano, Markus Reischl, and Pavel A. Levkin (2016) Evaluation of the Droplet-Microarray Platform for High-Throughput Screening of Suspension Cells. SLAS Technol. 22(2):163-175. doi: 10.1177/2211068216677204.
Ana I. Neto, Konstantin Demir, Anna A. Popova, Mariana B. Oliveira, João F. Mano, and Pavel A. Levkin (2016) Fabrication of Hydrogel Particles of Defined Shapes Using Superhydrophobic-Hydrophilic Micropatterns, Adv. Mater. DOI: 10.1002/adma.201602350
A. A. Popova, T. G. Hartanto, E. Schmitt and P. A. Levkin (2016) Droplet-Microarray on superhydrophobic-superhydrophilic patterns for high throughput live cell screenings RSC Adv (6): 38263-38276.
Popova A, Schillo S, Demir K, Ueda E, Nesterov-Mueller A, Levkin P (2015) Droplet-Array (DA) Sandwich Chip: A Versatile Platform for High-Throughput Cell Screening Based on Superhydrophobic–Superhydrophilic Micropatterning, Adv. Mater. 27: 5217–22
Nurgazieva D, Mickley A, Moganti K, Ming W, Ovsyi I, Popova A, Sachindra, Awad K, Wang N, Bieback K, Goerdt S, Kzhyshkowska J, Gratchev A (2015) TGF-β1, but not bone morphogenetic proteins, activates Smad1/5 pathway in primary human macrophages and induces expression of proatherogenic genes, J Immunol. 194 (2): 709-18
Popova A, Kzhyshkowska J, Nurgazieva D, Goerdt S, Gratchev A. (2011), Smurf2 regulates IL17BR by proteasomal degradation of its novel binding partner DAZAP2, Immunobiology 217 (3): 321-8
Popova A, Kzhyshkowska J, Nurgazieva D, Goerdt S, Gratchev A. (2010), Pro- and anti-inflammatory control of M-CSF-mediated macrophages differentiation, Immunobiology 216 (1-2): 164-72
Gratchev A, Kzhyshkowska J, Kannookadan S, Ochsenreiter M, Popova A, Yu X, Mamidi S, Stonehouse-Usselmann E, Muller-Molinet I, Gooi L, Goerdt S. (2008), Activation of TGF-beta-specific multistep gene expression program in mature macrophages requires glucocorticoid-mediated surface expression of TGF-beta receptor II, J. Immunol. 180 (10): 6553-65.